The future of measuring cancer
Guest Olivier Gevaert is an expert in multi-modal biomedical data modeling and recently developed new methods in the new science of “spatial transcriptomics” that is able to predict how cancer cells present spatially and will behave in the future.
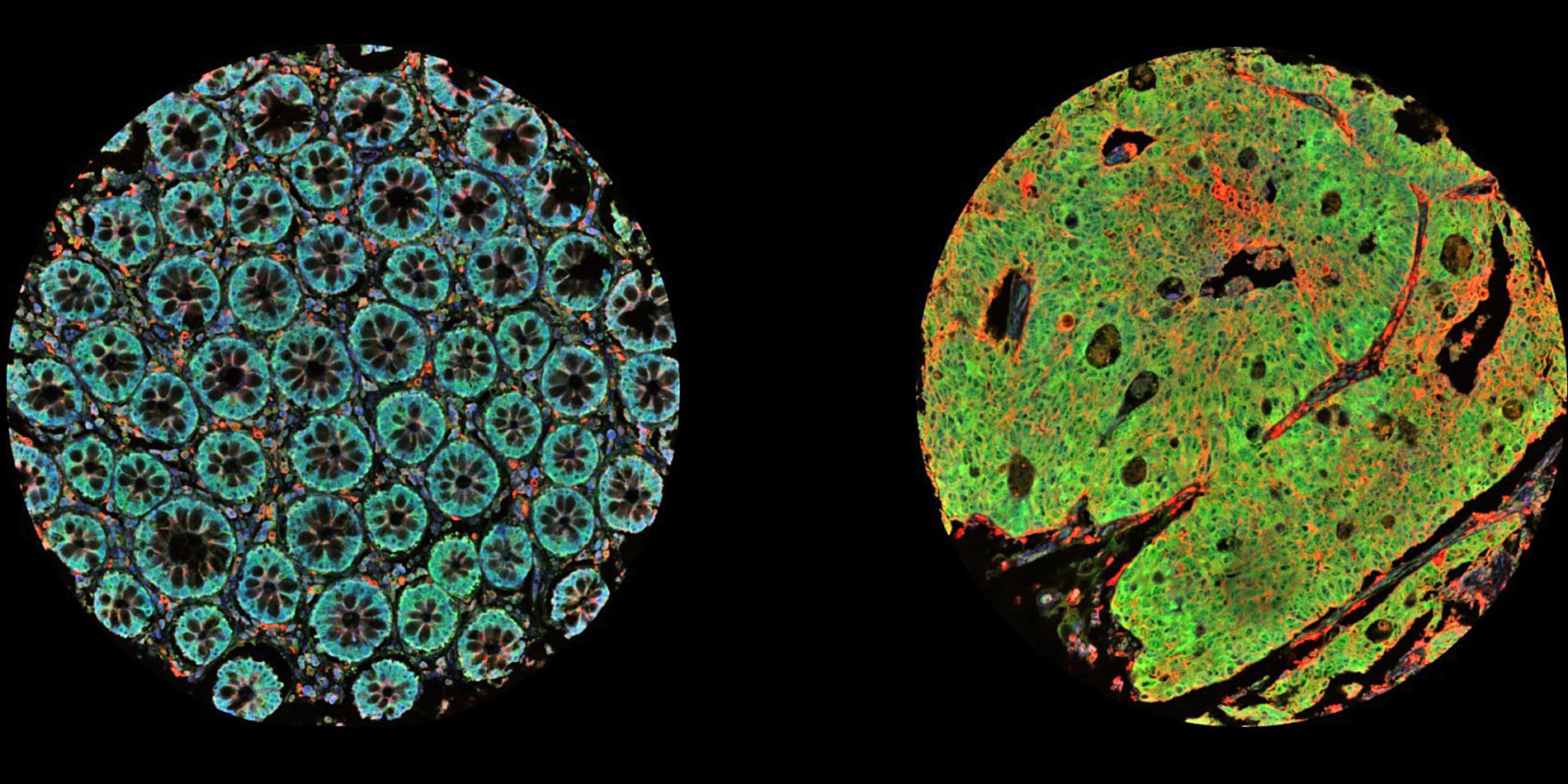
Tumors are not monolithic, he says, but made up of various cell types. Spatial transcriptomics measures cells in the undisturbed organization of the tumor itself and enables a more detailed study of tumors. This new technology can be used to determine what type of cells are present spatially and how each cell influences neighboring cells. It paints a picture of tumor heterogeneity, Gevaert tells host Russ Altman on this episode of Stanford Engineering’s The Future of Everything podcast.
Listen on your favorite podcast platform:
Transcript
[00:00:00] Olivier Gevaert: Now, with these spatial transcriptomics technologies, we can also capture the spatial information. Because tumors are very heterogeneous, there's lots of different cell types, there's intra and interpatient heterogeneity. And the spatial techniques allow us now to profile the transcriptome at every spot in a particular piece of tissue. And that gives us now more information about sort of which styles attract with each other, what pathways are active across sort of a cancellation.
[00:00:35] Russ Altman: This is Stanford Engineering's The Future of Everything podcast, and I'm your host, Russ Altman. If you enjoy the future of everything, please follow or subscribe wherever you get your podcasts. This will guarantee that you'll never miss a new episode.
[00:00:47] Today, Olivier Gevaert will tell us that there are new technologies, amazing technologies, for making measurements of cancer cells. These measurements at the molecular level are so good that you can use them to predict what the cells will look like and what they will behave like in the future. It's the future of measuring cancer.
[00:01:08] Before I get started, please remember to follow the podcast if you're not doing so already, and hit the bell icon if you're listening on Spotify. This ensures that you'll get alerted to all the new episodes, and you'll never miss the future of anything.
[00:01:27] You know, cancer continues to plague many patients across the world. And it's a cause of suffering, morbidity, and mortality. It's a complex disease. A single cancer is not just one type of cell, it's a cancerous population of cells. And not only are the cancer cells there, but immune cells, vascular cells, connective tissue cells, it's a mix of cells and they all kind of conspire to create this terrible disease.
[00:01:54] But, you know, there's new ways to make measurements of cells. One thing you can do is in any given cell, even though there are twenty thousand genes in the human genome, only about five or six thousand of them are active in each individual cell type. So when you look at a cell, if you can understand which of the genes are active and which are inactive. That gives you a ton of information about what's going on in that cell and maybe how you can kill it if it's a cancer cell.
[00:02:21] Well, Olivier Gevaert is a professor of Medicine, Biomedical Informatics, and Biomedical Data Science at Stanford University. And he's an expert at using these new cancer data types to analyze and understand cancer. Not only does he use one data type, he actually is an expert at taking several data types. The molecular data, the imaging data, the pathology data, and combining them all in interesting and important ways to understand the disease and how we might be able to develop new treatments.
[00:02:52] Olivier! A new method for studying cells is called spatial transcriptomics. So for those people who don't think about cancer and cells and measuring them, tell us a little bit about why there's so much excitement about spatial transcriptomics?
[00:03:07] Olivier Gevaert: So most previous technologies that allow us to profile the cells and tissues don't capture the spatial information in, uh, for example, cancer lesions. And they just grind up the tissue and then you can extract, for example, transcriptomic information, so expressions of all genes in the human genome over their activity from those tissues. And now with these spatial transcriptomics technologies, we can also capture the spatial information. Because tumors are very heterogeneous, there's lots of different cell types. There's intra and interpatient heterogeneity. And the spatial techniques allow us now to profile the transcriptome at every spot in a particular piece of tissue. And that gives us now more information about sort of which cells interact with each other, what pathways are active across a cancer lesion.
[00:04:01] Russ Altman: Okay, great. So let me just get, make sure I got this right. So I think it's generally understood that there are about twenty thousand human genes in the genome. But that every cell doesn't need all of them. So the cells turn on and turn off the genes that they need. And transcriptomics is the study of which genes, a transcript is a gene that's being turned on.
[00:04:21] Now, can you measure how many of these genes can you measure in the cells that you're looking at spatially? So you said that when you ground them up, I'm pretty sure you can measure all twenty thousand. But you don't have the information of which cell is doing which gene. So tell me a little bit more about the resolution and capabilities of this technology.
[00:04:41] Olivier Gevaert: So, as you said, there's about twenty thousand genes in the human genome. Uh, in normal sequencing approaches, we can, uh, capture all of them. In spatial, once we go down to, um, a bunch of cells, of course, not all these cells are good to express all these genes. So, typically, what we see is about four or five thousand transcripts being active in a single cell. So that's sort of what we're capturing with, uh, spatial transcriptomic technologies.
[00:05:07] Russ Altman: So you can measure all the active genes in the cell?
[00:05:10] Olivier Gevaert: Yes. Yep. So these techniques allow to capture all of them. So it's an unbiased, uh, technology. So you're not, um, uh, looking at only a portion of the transcriptome, but of course, once you go down to, uh, smaller groups of cells, not all genes are going to be active. So we're capturing the number of genes that are actually active or expressed is limited to a few thousand.
[00:05:34] Russ Altman: Okay, so now I'm going to ask you to give me a little tutorial on cancer because you said something very interesting, which was that, um, cancers are heterogeneous, they're made out of multiple cell types. And that might be a surprise to some people because when you think, uh, I have lung cancer, you just think, well, there are a bunch of lung cells that are proliferating too much and causing problems. Or when I have liver cancer, well, it's the liver cells, but it sounds like it's actually more complicated than that.
[00:05:59] So tell me what is the new way or the current way of thinking about these cancers and the types of cells that are involved? And why is it so important to understand these differences across the cells in a cancer?
[00:06:10] Olivier Gevaert: So first of all, the cancer cells themselves can evolve independently of each other, or they can, uh, there can be different clones in a particular tumor. For example, one tumor that's evolved over time, or maybe even in the context of treatment, right?
[00:06:24] So if you give them chemotherapy, maybe some, uh, certain cells are becoming resistant, so that creates a lot of heterogeneity within the cancer cells. And then there's also all other types of cells. There's immune cells, there's vascular cells, there's, uh, other types of cells that, uh, are normally present in tissues. And cancers, actually cancer or tumor actually learn how to, uh, either, um, uh, co-opt these cells and make them work for the lesion. Um, or these tumor cells, these immune cells can also attack cancer cells. And that's, for example, one of the reasons why immunotherapy has, uh, arisen, um, in the last, uh, couple of years, uh, as a treatment option for cancers. Because it uses actually your own immune cells to attack cancer cells.
[00:07:12] Russ Altman: Right.
[00:07:12] Olivier Gevaert: And yeah, with spatial technologies, now we actually have the opportunity to look at all these different, uh, interactions.
[00:07:19] Russ Altman: Great. So thank you. You've given us a little tutorial on cancer cells. Now we should think of it as a community of cells, not just one type of cell. And also you've given us this tutorial about this amazing new spatial ability to measure the genes.
[00:07:31] Well, I know that your group has recently written some really exciting papers about in the area of brain cancer, I believe. So tell us how this all comes together to really advance the field in terms of our ability to understand and maybe even treat these cancers.
[00:07:45] Olivier Gevaert: So, brain cancers are, especially glioblastoma, are very aggressive tumor type. There's not really a lot of treatment options for this disease. The survival is pretty short. And it's known that, in particular, glioblastoma is a very heterogeneous type of cancer. So lots of inter and intra tumor variability between patients and within patients. And so we use these spatial, uh, omics techniques to study brain tumors, uh, to learn more about sort of, uh, the spatial distribution of the different tumor cell types that are existing in these tumors and also the different immune cells that, uh, that are interacting with these tumor cells. Um, and that's sort of one, uh, yeah, application of this technology that we did recently.
[00:08:28] Russ Altman: Yeah, so what did you learn? I mean, so glioblastoma, as you said, is a dreaded disease. And, um, I guess you had some valuable samples probably from surgeries where they had taken the cancer out. Um, I'm sure there were surprises there.
[00:08:42] Olivier Gevaert: So we, first of all, we, uh, this is still a very expensive technology, right? Spatial transcriptomics. So we can't, uh, actually use this as a routine technique yet for studying these types of patients and for studying tissues. So we had, uh, spatial, uh, transcriptomic data of about twenty-five glioblastoma patients. And then we uh, essentially learned that certain cell types, uh, and also the relationship, the spatial relationship of these cell types, uh, predicts prognosis of these patients.
[00:09:13] Russ Altman: Oh.
[00:09:13] Olivier Gevaert: So that's kind of one of the main sort of takeaways of the study that we did.
[00:09:18] Russ Altman: Can you tell us anything about what were the differences, what leads to a more severe, uh, aggressive tumor and what leads to a tumor that's less aggressive?
[00:09:28] Olivier Gevaert: So first of all, we looked at all the spatial transcriptomic data of all these patients, right? So we have, let's say twenty-five patients, then we have these spatial transcript profiles up to, uh, four thousand, uh, transcript profiles spatially for each patient. So times the number of patients, we had, uh, roughly seventy-five thousand transcript profiles. And the first thing we did is we looked at all the different cell types that were present, uh, in these tumors.
[00:09:52] And we found about five cell types that also reflect sort of normal brain development and normal brains. And so then we looked at these five cell types and an examples, for example, uh, oligodendrocytes, uh, neural, um, cells, uh, astrocytes and mesenchymal cells. And these sort of are sort of normal cells that have turned malignant. And they represent sort of the heterogeneity in brain tumors. And then what we did is we looked at, um, how we, how these cells are spatially organized in these tumors. And we found that, for example, reactive astrocytes, uh, when they, uh, that they, uh, when they cluster with their own cell type, then it's very bad for prognosis.
[00:10:33] Russ Altman: Ah.
[00:10:34] Olivier Gevaert: But if those reactive astrocytes cluster with other cell type, for example, the mesenchymal cells or with, uh, with another cell type, then it's actually, uh, predictive of good prognosis. So that's sort of like one of the new types of analysis that we can do with spatial omics data. We can look at the spatial distribution.
[00:10:53] Russ Altman: Yeah. So that's great. And that actually leads to a very basic question that I didn't ask, which is how many cells, like, how big is the field of view that you're looking at? Because of course, when they cut out the cancer, it's macroscopically, it could be large. Are you looking at like, um, ten or twenty cells in a cluster or hundreds of cells or thousands of cells? Uh, I guess there's a decision you have to make about how broad of a field to look at.
[00:11:18] Olivier Gevaert: So each spot, uh, within, so we have a whole slide, which is a pathologist normally diagnoses a tumor based on a slide of tissue, which is put on a glass slide, and then they look at it under a microscope. And each spot which represents sort of a transcript profile that we can get from these tissues represents on average between ten to forty cells.
[00:11:40] Russ Altman: Okay.
[00:11:40] Olivier Gevaert: So it's not yet single cell, which is one thing that the field wants to move towards, but we get relatively down to a low number of cells. And then we actually use computational methods to, um, decouple all the signatures and make it single cell. So we actually can use computational algorithms to infer the expression of each individual cell in a particular spot.
[00:12:03] Russ Altman: That's exactly where I wanted to go next, which is, uh, everybody's been hearing about AI being applied to everything. And I wanted to ask what the role of AI and machine learning, if any, is in this kind of work.
[00:12:16] Olivier Gevaert: So that's the kind of one of the first things that we did is we can use AI and machine learning to, um, make the resolution of the data even, uh, smaller, right? So in a sense where we can get, uh, estimates of what each individual cell is doing in each of these, uh, spots. And then the second step is that, um, so I'd mentioned before that these spatial omics technologies are very expensive, right? We can actually not apply them on a large scale. So then we used actually an AI method to map these cell types that we discover on what we call, uh, these whole slide images. Uh, so we use images to predict, uh, these cell type distributions across a slide, for example. So then we don't actually have to go collect the spatial task atomic data.
[00:13:04] Russ Altman: Ah, and good. So I wanted to get into that too, cause I know that you've been doing a lot of work in this area called whole slide imaging and whole slide. I believe that you're using these images with genetic and genomic and transcriptomic data. So tell me a little bit about what that challenge is like, uh, you know, a whole slide is literally like you said, a pathologist cuts up the sample, puts it on a glass slide, that's the whole thing. And so now how are you developing methods to kind of improve our ability to analyze those slides? It's not just the eyeballs of the pathologist anymore, is it?
[00:13:37] Olivier Gevaert: Yes, that's correct. Yeah, so you have that whole image. It's a very high-resolution image. It's a few hundred thousand by a few hundred thousand pixels. And we essentially use all the recent AI and deep learning methods that, um, uh, all these images to then make predictions. Uh, and so with the spatial omics technology, now we have this whole slide and we have for each, um, uh, spot in that slide, we have a whole transcript profile. And from that transcript profile, we predict the cell types, and now we can predict those cell types from the image.
[00:14:07] Russ Altman: Ah.
[00:14:08] Olivier Gevaert: And then these images, these whole celled images, this is sort of the routine, uh, technology that's being used across all, across the world to diagnose cancer patients, not just brain tumour patients. So now we can roll out this model and make predictions across larger cores. And that's also what we did in this study. We went to a big project, which is the Cancer Genome Atlas, which has, uh, for a few hundred, uh, brain tumor patients has these whole slides. And now we can predict these cell types and, uh, look at the spatial distribution of these cell types, but in much larger cohorts, which allows us to then correlate it with treatment, with prognosis, um, and get more reliable correlations.
[00:14:49] Russ Altman: So, yeah, so let me just make sure I have this right. So based on your spatial genomics, you can get a good idea of like a region, a small region of cells, which the cell types are, what they look like, because you also have a picture of them, and you can understand the genes that are turned on or turned off. When you do that for a small set of cells, though, that gives you enough information to then look at this big slide, which I'm guessing has thousands if not millions of cells and based on how much they look like the cells that you've just analyzed in detail you can make a guess for what they're, what's going on inside those cells biologically. And so kind of for free, of course, it's a ton of work for you, I understand that, but kind of for free, I now have my whole cell labeled as, okay there's a bunch of this type of cell and there's a bunch of this type of cell and here's where they are. And then you can start looking at what does that mean for prognosis or options for treatment. Does any of this, uh, make any implications early on for treatment or is that still far away?
[00:15:44] Olivier Gevaert: For treatment, of course, there's ways of, um, maybe informing drug discovery, right? Because by being able to make these predictions, we know which cell types are close to each other and how it is correlated with prognosis. And this could inform drug developers, uh, in terms of which cells are aggressive, uh, which cells do we need to target. Then what I said before is that certain um, cells, if they, uh, cell types, if they cluster together, it's, uh, related to good prognosis. So perhaps we need to, uh, develop drugs that can attract these cell types to the tumor, like immune cells or other types of cells, because then it could actually, uh, make the tumor less aggressive.
[00:16:27] Russ Altman: Great. How do the pathologists feel about all this? So I could imagine that they're excited because it's going to improve their work, or I could imagine that they feel like they're losing control because after training and doing things a certain way with the microscope and with the slides, now you're coming with these very disruptive ways of looking at. So tell me a little bit about what that dynamic is like with the practitioners.
[00:16:48] Olivier Gevaert: So the type of models that we develop on these whole slides actually provides extra information to the pathologists that they actually can't, um, perceive themselves, right? We are actually using the images as a surrogate to reflect molecular properties, uh, with these spatial, uh, profiles. So we actually are providing them with additional information that then they can use to maybe come up with a better, uh, understanding of, um, of the diagnosis of disease. And perhaps also, I mentioned before, these images are huge, right?
[00:17:20] Russ Altman: Yeah.
[00:17:20] Olivier Gevaert: There's hundred thousands by hundred thousands of pixels. They don't always have time to look at every aspect or every region in the image. And with these tools that we developed, we can point them to the right areas. We can look at more aggressive areas that are predicted by our model, um, maybe different types of, um, areas that have certain cell types. And it can also see, um, predictions of, for example, certain pathways that are being activated. I mean, it could then also help with, for the medical oncologists, uh, with treatment decisions.
[00:17:54] Russ Altman: Right. And I'm also guessing that, I know that pathologists often have what they call these staging or ratings where they say, based on what I'm seeing, I'm going to give it a one, two, three, or four based on how aggressive it is or whatever. I'm guessing that this new information may in the future change our ability to have fine grain staging information. Is that likely?
[00:18:15] Olivier Gevaert: Um, sure. I think it, well, it can definitely help with better staging. Also, maybe one other aspect is that we can more democratize the pathology knowledge, right? Because maybe at Stanford, we have, uh, expert pathologists, but maybe in, um, in other areas with less expertise or with less bandwidth, we can now sort of capture this knowledge in these AI models and then use it to parse a world.
[00:18:40] Russ Altman: This is The Future of Everything with Russ Altman. We'll have more with Olivier Gevaert next.
[00:18:57] Welcome back to The Future of Everything. I'm Russ Altman, and I'm speaking with Professor Olivier Gevaert from Stanford University.
[00:19:03] In the last segment, Olivier told us about this amazing new ability to measure cells as they sit in a cancer, interacting with one another at a molecular level. That's giving us an amazing ability to see what cells are there and what's wrong with them in cancer.
[00:19:19] In the next segment, he's going to tell us that it's not just about those measurements. There are multiple measurements that need to be combined to build the best models for the cancer cells and how to kill them. He'll also tell us that, in a funny way, synthetic data, fake data, generated from real data, is incredibly important for this pursuit. And he'll end with a vision of digital twins. Copies of your cancer in a computer, where we can treat it in the computer before we treat you with your cancer. That should lead to more effective strategies for treatment.
[00:19:52] So Olivier, one of the things that I know you do a lot of is what they call, is what you call, multimodal kind of data fusion. Uh, a lot of words there. Can you break it down for me? What is the challenge and what is the opportunity for a multimodal data?
[00:20:07] Olivier Gevaert: So, for complex diseases like cancer, we believe that we need to harness sort of all the different data modalities that are being collected routinely from patients. So we have some molecular information that's routinely collected, like mutations, maybe RNA sequencing, although RNA sequencing is still mostly a research tool. Then digital pathology, these whole slide images are always routinely collected because they're sort of used for staging and diagnosis of disease. And then we also have radiology, uh, CT, MRI, the images, um, which are also non-invasive modality, right? So that's a modality that's also used during treatment to track, um, the progress, uh, whether tumors are recurring. Um, and so we believe that's by combining all these different types of information, we can use them to make predictions about treatment, about diagnosis, and sort of realize this idea of precision medicine. Uh, especially in the context of like complex cases where maybe we cannot fully explain treatment response.
[00:21:11] Russ Altman: Okay. So tell me a little bit about how you would even start to take, I don't know, molecular data and combine it with like imaging data. Like, yes, I can see how it's relevant. But it sounds like it might be two separate analyses. But I think you're suggesting that there might be a unified analysis that's more powerful.
[00:21:30] Olivier Gevaert: Yeah. So we designed these AI machine learning approaches that can take all these different data modalities, these transcript profiles, then these large 2D, uh, digital pathology images. And then we have 3D radiographic images. And what we do is we learn sort of these representations of all these different modalities using deep learning methods and represent them in sort of a lower dimensional space. And then we actually create sort of these vectors that represent these data modalities. And then we can sort of combine them. Uh, in, uh, machine learning methods, I make predictions about the future or about treatment.
[00:22:07] Russ Altman: Okay, so what are the predictions that you're making? Is it about the aggressiveness of the tumor or the likelihood of responding to drugs or all of the above?
[00:22:15] Olivier Gevaert: First, the most common, uh, outcome we have from patients is overall survival. So we know when patients die. Um, which is a more distant outcome is not always the best outcome to learn from. But that gives us a clue of, uh, what features are relevant. And then in more in cohorts where we have more detailed information about sort of the first line of treatment that's being used on these patients, we look exactly at treatment response.
[00:22:38] Russ Altman: Gotcha.
[00:22:38] Olivier Gevaert: The other thing that we can do is we can use the radiology images to track the tumors and track the tumor growth. And then we can actually use that size or how the tumors are growing or shrinking as a way of, uh, as an outcome and we can try to predict that.
[00:22:54] Russ Altman: So I'm fascinated because in your brief description just now about how you use the AI, it clearly, it became clear that this very quickly turns into a mathematical abstraction. You were talking about vectors and mathematical vectors and AI and machine learning. How do you, and as you know very well, one of the issues about AI these days is what they call the explain-ability or the interpretability. The need for the user of the AI to kind of understand what the AI did. Because at the end of the day, the physician is the one who's going to make a decision. And if the physician is relying on AI that they don't understand, it could be a problem.
[00:23:29] So how is your group kind of dealing with that issue of taking these complex mathematical models and turning them into something that a well-trained doctor who's not a mathematician can understand.
[00:23:42] Olivier Gevaert: So, interpretability of these deep learning methods is hard in and of itself. And then when you bring it into a multi modal context, it becomes harder.
[00:23:50] Russ Altman: Yes.
[00:23:51] Olivier Gevaert: But we try, we still try to, uh, make sense of these models. First of all, we can try to figure out which components, which modality is helping the most in terms of making a prediction.
[00:24:02] Russ Altman: Ah.
[00:24:03] Olivier Gevaert: So that can give us some clue of is it the transcript profiles? Is it maybe the radiology image? Is it the size of the tumor? Like which, which feature it is actually making the biggest contribution. The other thing we can do is the image modalities. We can sort of propagate back the signal and make visualizations in these images of what areas of the image are the most important in making the prediction.
[00:24:27] Russ Altman: Like what was the AI paying attention to?
[00:24:30] Olivier Gevaert: Yeah, yeah, exactly. Exactly. For example, in the radiographic image, we could look at like, which is it, uh, the tumor itself? Is it the environment where the tumor is located? In pathology images, what cells are in areas that are important? So we can get some clues about sort of why is the model making these, uh, these predictions?
[00:24:49] Russ Altman: Yeah, I could imagine for the pathology, it might be very exciting for a pathologist to look at a slide and then have you create, you know, little, uh, apps that like light up the cells that are getting your AI very nervous, and that could draw their attention to these cells. And either they would agree and say, yep, I can totally see that, or they might be surprised. But either way, they've learned something and that they can integrate in.
[00:25:12] Well, I wanted to move to this idea. You touched upon it in the first segment of synthetic data. So, um, we all know that AI requires lots of data and sometimes even in medical situations, it seems like a lot of data to us. But it's not quite enough to feed the AI.
[00:25:29] So what's the role of synthetic data in your work?
[00:25:31] Olivier Gevaert: So in this, one of the main challenges with multimodal modeling is that we don't always have all these modalities for all patients. So, it's hard to get cohorts that are big enough, um where we have all these different modalities. So one of the things that we have, um, done in our work is we've, uh, developed these models that can predict one modality from another.
[00:25:52] For example, we can, uh, use digital photology images and we predict transcript profiles. And we also did the opposite. We have sort of transcript profiles and we generate synthetic images, uh, that reflect sort of the…
[00:26:03] Russ Altman: So you, that sounds cool. So you're taking the, which genes are turned on and off and then creating pictures of cells, like interacting with one another?
[00:26:11] Olivier Gevaert: Exactly. Yeah. And we can do this across tissues. Uh, and it reflects of the cell types that are present in these transcript profiles. And we've, uh, shown these pictures, uh, to pathologists that they cannot distinguish the synthetic images from the real ones.
[00:26:25] Russ Altman: Wow. Wow. So this is like dolly for cells.
[00:26:29] Olivier Gevaert: It's exactly like that. So we encode the transcript profiles as text, and then the, we generate sort of the synthetic images based on those gene profiles.
[00:26:39] Russ Altman: Fantastic. So, and this then you can use to fill out your data set, as you said, it would otherwise be missing. But now you can include that patient because they not only have the data that was measured, but you've synthesized the other data that's good enough. So that you can present it to the AI for learning and for classification. Fantastic.
[00:26:58] Okay, the final thing in the last couple of minutes, I wanted to ask you a little bit about this idea of digital twins. It's been in the air. Uh, you know, the general, I think the general idea is that you have data about a real patient and you create a model of that patient in the computer, where you can do a lot more things to the model, like give them drugs, watch them over time, maybe accelerate time so you can watch, see it happen faster.
[00:27:22] What is the role of these digital twins in your work? What's the vision?
[00:27:26] Olivier Gevaert: So that's sort of with the work that we are doing in synthetic data, multimodal modeling, that's the goal that we're working towards. We want to develop, for example, a digital twin of a lung cancer patient, where exactly we can do what you're describing. It's sort of a virtual representation of that tumor lesion. We want to this model to be patient specific and that we can run experiments on this model. We can run different treatment strategies so we can, uh, look at simulations of the outcome and then figure out what treatment this patient needs.
[00:27:57] Russ Altman: Great. And where are we in that? So it sounds like the synthetic data is a little bit in that direction because you're already synthesizing. I think you said the transcriptomic profile from the cell pictures or the cell pictures from the transcriptomic. So in a sense, that's already testing your ability to make, uh, faithful models. Because then you could always just then look at the biopsy and say, does it match the picture that I generated synthetically? And if they look pretty similar, that means you're doing a good job at your synthetic, that aspect of your digital twin.
[00:28:30] Olivier Gevaert: Yeah, that's exactly right. So the work we did on digital pathology, we're also, we have some preliminary work in the context of radiology where we can start manipulating, uh, and creating sort of synthetic images of these tumors and how they look in, uh, radiology images. And we can sort of shrink and grow these lesions with our machinery models that we've built. And now sort of the next step is, I mentioned before that, uh, radiology, like CT images of lung cancer patients, uh, or CT imaging is being used to track patients over time.
[00:29:01] So we've all these growth curves. We want to model them now and then we want to create sort of these models that are capable of, uh, inferring these trajectories. And that sort of would put us on a path to developing a digital twin.
[00:29:16] Russ Altman: Thanks to Olivier Gevaert. That was The Future of Measuring Cancer. Thanks for tuning into this episode.
[00:29:23] With close to 250 episodes in our back catalog, you have a wide array of discussions on the future of many things at your disposal and instantly. If you're enjoying the podcast, please consider leaving a five-star review and comments to tell people why you love it so much. Your feedback helps grow the show and helps other people discover it.
[00:29:44] You can connect with me on X or Twitter @RBAltman, and you can connect with Stanford engineering @StanfordENG.